There are several different ways to figure out your genetic ancestry. One way that 23andme shows your ancestry is by comparison with reference populations of the HGDP (Human Genome Diversity Project) dataset. I have listed how similar I am to various groups in the table below:
| Reference Population | Similarity | Groups Included |
|---|---|---|
| Central & South Asians | 67.14 | Pathan, Makrani, Kalash, Hazara, Balochi, Sindhi, Brahui and Burusho |
| Northern Europeans | 66.94 | western Russia, France, Orkney Islands |
| Southern Europeans | 66.93 | northern Italy, Tuscany, Sardinia, French Basque |
| Near Easterners | 66.82 | Palestinian, Druze, Bedouin |
| Siberians | 66.55 | Yakut |
| Eastern Asians | 66.48 | Japan, Cambodia, China (Dai, Daur, Han, Hezhen, Lahu, Miaozu, Mongola, Naxi, Oroqen, She, Tu, Tujia, Uygu, Xibo, Yizu) |
| North Americans | 66.47 | Pima, Maya |
| South Americans | 66.43 | Surui, Karitiana, Piapoco, Curripaco |
| Oceanians | 66.38 | Papuans, Melanesians |
| Northern Africans | 66.16 | Mozabite |
| Eastern Africans | 64.13 | Kenya |
| Southern Africans | 64.04 | San, Bantu speaking South Africans |
| Central Africans | 64.01 | Biaka, Mbuti Pygmies |
| Western Africans | 63.98 | Mandenka, Yoruba |
My numbers are not too different from anyone from the northwestern part of the South Asian subcontinent.
One thing to consider over here is that you are being compared to a specific set of populations. As you can see, there is no Indian references here. Similarly, Near Easterners are represented only by samples from Israel and North Africans by one Algerian population. I wonder what the case would be if they had Egyptians or Ethiopians etc in their reference.
Another way to look at your genetic ancestry is with a PCA (Principal Component Analysis) plot. With the same reference populations mentioned above, 23andme calculated the two dimensions of largest variation among that data. These two axes don’t completely describe the variation across the samples, but being the two largest components they can be used to project your genetic data in that space. At the world level, I am the green marker in the middle of the Central/South Asian cluster.
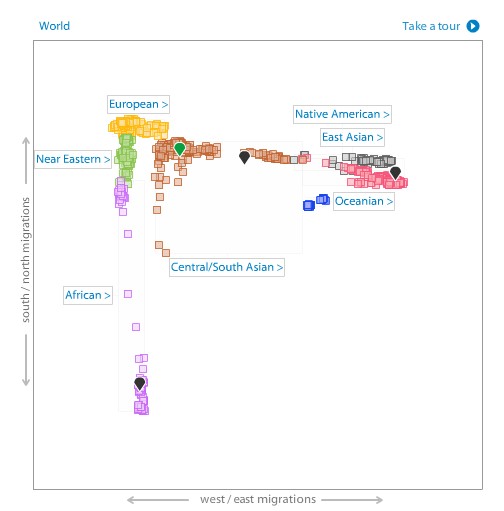
In the South Asian PCA plot, I am in the middle of the Pathan cluster and right at the top edge of the Sindhi one.
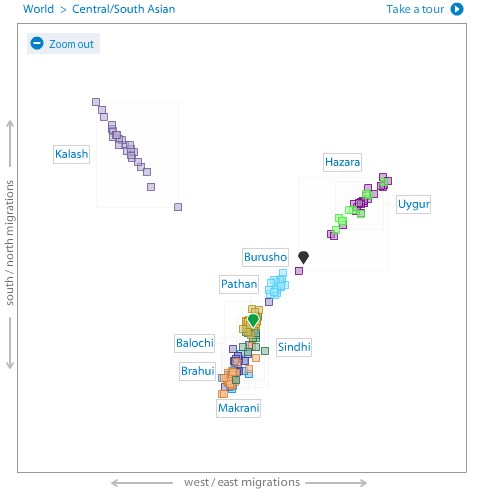
Now this doesn’t make me a Pathan. For one thing, 23andme’s reference populations do not have any Punjabis. I am sharing with a number of North Indians and Pakistanis, including several Punjabis, and they all lie around me in the plot.
There is another problem with a PCA plot though. We are looking at the two most significant dimensions, but there are other dimensions too and they combined together could account for a lot of the variation among people’s genomes. Also, let’s say we have someone who is a child of a European and an East Asian parent. Now that person, who is 50% East Asian and 50% European, would be placed about midway between the East Asian and European clusters. That’s where the Uygur and Hazara clusters are. So we can’t say that someone is Uygur just because they are placed in the Uygur cluster in a PCA plot.
There are other ways to look at your genetic ancestry and I have been exploring a bunch of them. We’ll talk about them next.
hat’s where the Uygur and Hazara clusters are. So we can’t say that someone is Uygur just because they are placed in the Uygur cluster in a PCA plot.
yeah, my family is on the edge of the ughyr/hazaras in the global. just an artifact of the biased samples and low dimensionality.
also, me
67.66 east asians
67.47 siberians
67.39 oceanians
67.33 s. americans
67.20 n. americans
67.18 c/s. asians
66.56 n. europeans
66.47 s. europeans
66.38 n. easterner
65.78 n. african
64.15 n. african
64.04 s. african
64 c. africans
63.99 w. africans
Interesting that you are closest to East Asians, though it makes sense considering their population groups.
Also your similarity measures are much higher than mine. Even your C/S Asian one (#6 on your list) is higher than mine (#1 on my list). But that’s old news now.